Latest Research
- 2019.03.01
- Ueda-Kitaguchi Group
- Ueda-Kitaguchi Group
- Ueda-Kitaguchi Group
- Ueda-Kitaguchi Group
- Ueda-Kitaguchi Group
- Ueda-Kitaguchi Group
- Ueda-Kitaguchi Group
- Ueda-Kitaguchi Group
- Ueda-Kitaguchi Group
- Ueda-Kitaguchi Group
"Luminescent detection of Protein-Protein Interaction by tagging tiny peptides"
It has been predicted that there are several hundred thousand pairs of protein-protein interactions in one cells of human. As protein-protein interactions play major roles for life phenomenon, understanding of protein-protein interaction is very important for various fields such as basic biology, diagnosis and drug discovery. In protein-protein interaction assay fluorescent proteins and luciferase enzymes often fused to interacting pair. However, the rather large sizes of the fluorescent proteins and the luciferase enzymes sometimes cause the problems of the misfolding and the steric hindrance. Therefore, in the present study, we developed a very small probe for protein-protein interaction assay1).
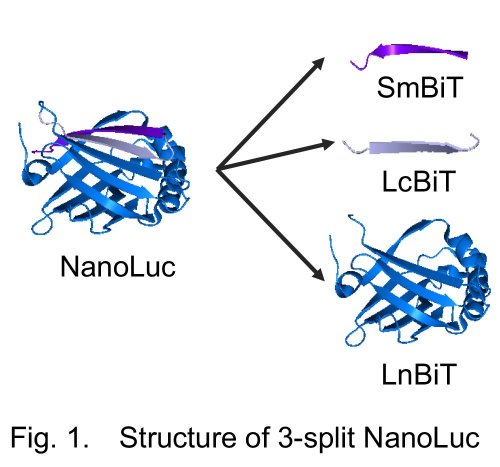
For the assay, we utilized a recently developed small luciferase "NanoLuc" originated from a deep sea shrimp. The luminescence of NanoLuc is very bright, and while its structure is very stable. NanoLuc is consisted of 11 β-strands. In a frequently used intracellular protein-protein interaction assay Protein-fragment Complementation Assay (PCA), a sensor enzyme or a fluorescent protein is divided into two fragments. In conventional PCA using NanoLuc, NanoLuc was divided into the C-terminal β-strand named "SmBiT" and the remaining part "LgBiT" 2). This time, we divided it into three, namely, one strand named "LcBiT (the C-terminal strand of LgBiT)", another strand "SmBiT", and the remaining part named "LnBiT (the N-terminal part of LgBiT)" (Fig. 1). Both LcBiT and SmBiT have fairly small sizes (11 aa ~1.2 kD).
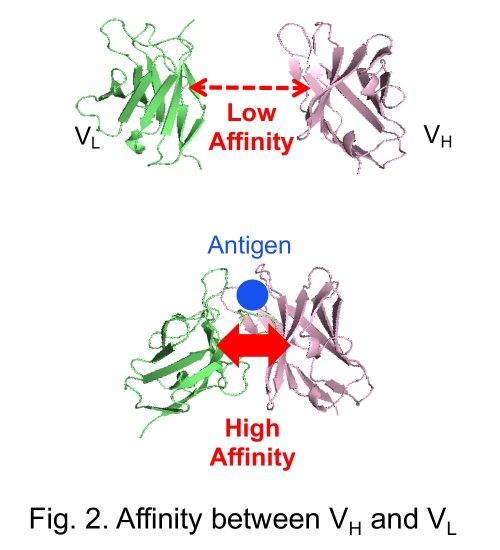
In this study, a pair of two antibody fragments, VH and VL that consist an antigen binding sites, was applied as an interacting pair. The affinity between VH and VL isolated from an antibody, especially when its antigen is a small molecule, is usually low in the absence of antigen. However, the affinity increases when antigen is added (Fig. 2). The antigen-dependent interaction between VH and VL could not be detected using the conventional NanoLuc PCA, probably due to the problem of folding and/or steric hindrance. Hence, we tried to assay the affinity by our new system using the two small peptides.
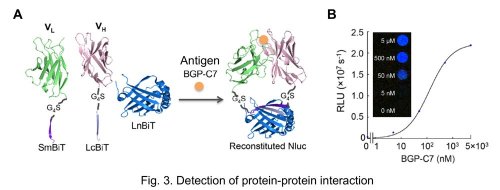
To this end, the VH and VL fragments were fused to LcBiT and SmBiT, respectively, expressed in E. coli and purified. LnBiT and antigen (osteocalcin peptide BGP-C7) was added to the mixture of VH-LcBiT and VL-SmBiT. As a result, the luminescent intensity significantly increased depending on antigen (Fig. 3A). Furthermore, the peptide sequence of SmBiT was optimized by taking a report measuring the affinity between SmBiT variants and LgBiT 2), into account. The KD value of SmBiT is 1.9×10-4 M, and those of the SmBiT variants applied to our test were 2.5×10-6 M, 1.8×10-7 M, and 0.7×10-9 M. As a result, when the SmBiT variant having the highest affinity was used, the signal was enhanced near 300-fold, while the signal/background ratio was maintained. Furthermore, the signal could be detected by a naked eye (Fig. 3B).
The interactions of proteins having complex structures were often undetected by conventional assays due to the probe size. This technology "NanoLuc ternary technology" could be a tool to analyze the interaction of such proteins. Now we are improving this technology to develop probes that can detect many more protein-protein interactions.
Reference
- 1) Y Ohmuro-Matsuyama & H Ueda, Homogeneous noncompetitive luminescent immunodetection of small molecules by ternary protein fragment complementation. Anal. Chem. 90, 3001-3004 (2018)
- 2) AS Dixon et al. NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem. Biol., 11, 400-408 (2016)