Latest Research
- 2019.10.01
- Tanaka-Yoshida Group
Establishment of modification technology for multiple nuclear genome loci in Cyanidioschyzon merolae, a model plant cell
[Research Background]
To resolve the complexity of living organisms, making use of simpler model organism has been an important strategy to shed light on the fundamental architecture. To this end, we introduced a primitive red alga Cyanidioschyzon merolae as a model plant cell to study a variety of basic aspects of photosynthetic eukaryotic cells. C. merolae contains only one chloroplast, one mitochondrion and one nucleus in a cell, and the completely determined nuclear genome sequence can be easily modified by homologous recombination[1].
Genome modification is a method to specifically change a specific locus of the genome, and this method usually requires a use of a selectable marker gene. Because the once used marker gene is usually left on the genome, the same marker gene cannot be used again as the marker gene for the second round of genome modification. Thus far, only two types of selectable marker gene are available in C. merolae, URA5.3[2,3] and CAT[4], and limitation of selectable marker genes has been restricting the utility of genome modification in C. merolae. In this study, we have developed a marker gene recycling system for repeated nuclear genome modification in C. merolae.
[Results]
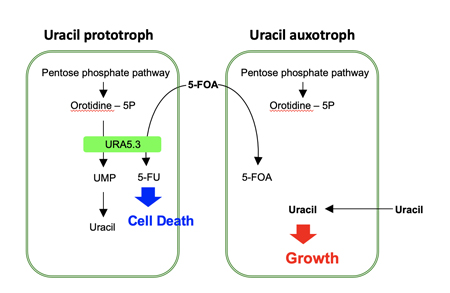
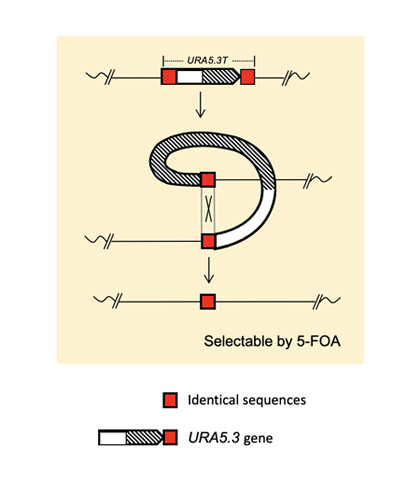
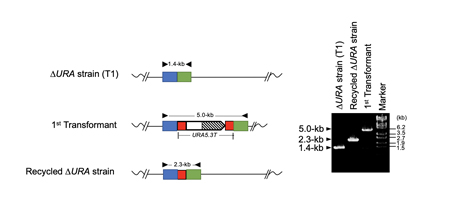
If a once used marker gene can be eliminated from the genome, it is possible to use the same marker gene for the second-round genome modification. For this purpose, we used the URA5.3 gene, encoding a fusion protein of orotate phosphoribosyltransferase and orotidine-5'-phosphate decarboxylase, as the counter-selectable marker gene. URA5.3 is an enzyme for the biosynthesis of uridine-5'-monophosphate (UMP) from orotidine-5'-monophosphate, however, the same enzyme converts 5-fluoroorotic acid (5-FOA) to toxic 5-fluorouridine (5-FU) (Fig. 1). Therefore, using a uracil prototroph cell as the parental strain, it is possible to select URA5.3 deficient cells on gellan gum plates containing uracil and 5-FOA. In this study, we designed a modified URA5.3 marker gene, URA5.3T, and constructed[5], where the URA5.3 open reading frame region was sandwiched by 500-bp identical DNA sequence corresponding to the URA5.3 terminator. After isolating a URA5.3T-genomic insertion mutant by homologous recombination, this URA5.3T marker gene can be eliminated by the 5-FOA selection through a homologous recombination mediated by the 500-bp repeated sequences on the both sides (Fig. 2). These recombination events were successfully confirmed by PCR analyses as shown in Fig. 3.
[Future perspective]
It is frequently found that one genome contains multiple homologous genes sharing redundant functions. In these cases, it is difficult to understand the function of these genes by knocking out one gene, and disruption of multiple genes would be essential to make clear the function. In this study, we have made it possible to modify multiple nuclear genes one by one in C. merolae, which is very useful a tool for molecular genetics analysis as well as for various biotechnological applications[6,7].
Fig. 1. Scheme of the effect of 5-FU (5-fluorouridine) for the cell growth and death
Fig. 2. Structure and homologous recombination by the URA5.3T marker gene
Fig. 3. Confirmation of recombination events by URA5.3T
[References]
- [1] Kuroiwa T. (1998) The primitive red algae Cyanidium caldarium and Cyanidioschyzon merolae as model system for investigating the dividing apparatus of mitochondria and plastids. Bioessays 20, 344-354.
- [2] Ohnuma M., Yokoyama T., Inouye T., Sekine Y. and Tanaka K. (2008) Polyethylene glycol (PEG)-mediated transient gene expression in a red alga, Cyanidioschyzon merolae 10D. Plant Cell Physiol., 49, 117-120.
- [3] Taki K., Sone T., Kobayashi Y., Watanabe S., Imamura S. and Tanaka K. (2015) Construction of a URA5.3 deletion strain of the unicellular red alga Cyanidioschyzon merolae: A backgroundless host strain for transformation experiments. J. Gen. Appl. Microbiol., 61, 211-214.
- [4] Fujiwara T., Ohnuma M., Kuroiwa T., Ohbayashi R., Hirooka S. and Miyagisima S.Y. (2017) Development of a double nuclear gene-targeting method by two-step transformation based on a newly established chloramphenicol-selection system in the red alga Cyanidioschyzon merolae. Front. Plant Sci., 8, 343.
- [5] Takemura T., Imamura S., Kobayashi Y. and Tanaka K. (2018) Construction a selectable marker recycling system and the use in epitope tagging of multiple nuclear genes in the unicellular red alga Cyanidioschyzon merolae. Plant Cell Physiol., 59, 2308-2316.
- [6] Yamaguchi S., Kawada Y., Yuge H., Tanaka K. and Imamura S. (2017) Development of new carbon resource: Production of important chemicals from algal residue. Sci. Reports, 7, 855.
- [7] Takemura T., Imamura S. and Tanaka K. (2019) Identification of a chloroplast fatty acid exporter protein, CmFAX1, and triacylglycerol accumulation by its overexpression in the unicellular red alga Cyanidioschyzon merolae. Algal Res., 38, 101396.