Latest Research
- 2021.07.01
- Tanaka-Yoshida Group
Evolutionary changes in DnaA-dependent chromosomal replication in cyanobacteria
Precise chromosomal DNA replication is required for the inheritance of genetic information during cellular proliferation. Chromosome replication is tightly controlled, mainly at the initiation stage of DNA replication. In most bacteria with a single circular chromosome, chromosome replication is initiated at a unique origin named oriC. oriC comprises several copies of the DnaA-box that is bound by DnaA. DnaA unwinds the duplex DNA to form single-stranded DNA templates. Subsequently, the replisome is recruited to the unwound DNA and then initiates DNA synthesis (1). Free-living bacteria, which do not possess dnaA, were not reported, and evidence suggested that the DnaA-oriC-dependent mechanism of chromosomal DNA replication initiation is conserved among bacteria, except for certain symbionts and parasites, which do not harbor dnaA. Moreover, dnaA is not encoded by the genomes of symbiotic cyanobacterial species and chloroplasts (2), which evolved from a cyanobacterial endosymbiont. However, the dependence of chromosome replication on DnaA-oriC in cyanobacteria other than S. elongatus, Synechocystis sp. PCC 6803, and Nostoc sp. PCC 7120 and the evolutionary relationships among DnaA-oriC-dependent and independent species have not been examined (2). Here, we introduce that loss of DnaA-oriC-dependency independently occurred multiple times during cyanobacterial evolution (3).
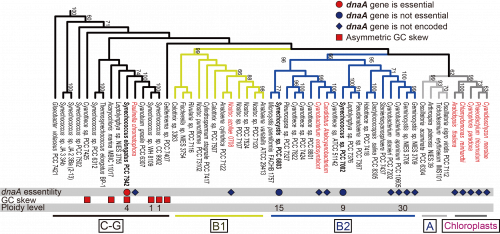
We first re-examined the distribution of dnaA sequences in these completely sequenced cyanobacterial and chloroplast genomes (Fig. 1). Consistent with our previous search (2), we did not detect dnaA in the genomes of symbiotic cyanobacteria (Fig. 1). The phylogenetic distributions of these symbionts and chloroplasts suggest that loss of dnaA independently occurred in their respective ancestors (Fig. 1). Although it has been believed that dnaA is conserved among free-living bacteria (1), our search revealed that the genomes of the free-living cyanobacterial species Cyanobacterium stanieri PCC 7202, Cyanobacterium sp. PCC 10605, Geminocystis sp. PCC 6308, and Geminocystis NIES-3708 and 3709 do not encode dnaA. These cyanobacteria formed a monophyletic group (Fig. 1). Further, the topology of the phylogenetic tree suggests that the common ancestor of this group (Fig. 1) lost dnaA independently from the ancestors of the above-mentioned symbiotic species and chloroplasts.
 |
|||
| Fig. 1. |
Phylogenetic distributions of dnaA among cyanobacterial and chloroplast genomes |
||
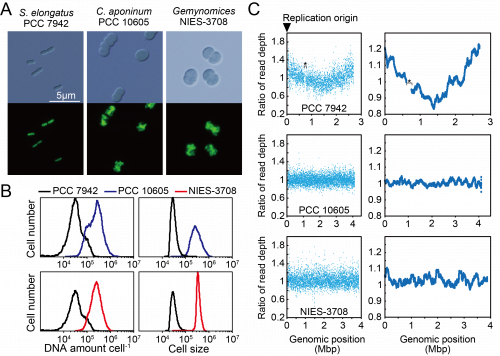
To determine the ploidy of C. aponinum and Geminocystis sp., the DNA levels and cell sizes were examined using flow cytometry (Fig. 2A and B). The fluorescence intensity of SYBR Green per C. aponinum and Geminocystis sp. were approximately 8-times higher compared with that of S. elongatus (Fig. 2B). Thus, the result indicates that C. aponinum and Geminocystis sp. possess approximately 30 copies of its chromosomes. Further, the volume of C. aponinum and Geminocystis sp. cells were approximately 8-times higher compared with that of S. elongatus (Fig. 2B), showing that an increase in chromosomal ploidy correlated with an increase in cell size.
Next, we examined where chromosomal replication starts by a DnaA-independent mechanism. When high-throughput sequence reads of exponentially growing C. aponinum and Geminocystis sp. were mapped to the reference genome, the read depth normalized by that of stationary phase cells was approximately constant throughout the chromosome (Fig. 2C). These results suggest that in the dnaA-negative cyanobacteria C. aponinum and Geminocystis sp., replication of multiple copies of the same chromosomes is asynchronously initiated from multiple sites rather than a unique point, as for Synechocystis sp. PCC 6803.
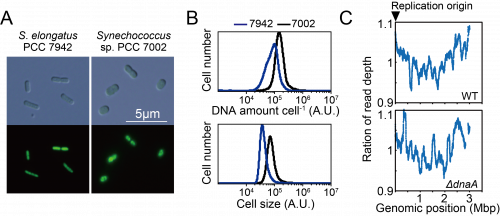
To examine the dependence of DNA replication on DnaA in Synechococcus sp. PCC 7002, which is phylogenetically closely related to dnaA-lacking free-living species as well as to dnaA-encoding but DnaA-oriC-independent Synechocystis sp. PCC 6803 (Fig. 1), we constructed a dnaA deletion mutant. When the high-throughput sequence reads of exponentially growing wild-type Synechococcus 7002 were mapped to the reference genome, a V-shaped profile was observed (Fig. 3C). The peak was detected around dnaA at position 1 (Fig. 3C), suggesting that the wild type possessed a unique replication origin near dnaA. When the sequence reads of the ΔdnaA clones were mapped to the reference genome, all clones exhibited a V-shaped profile similar to that of the wild type (Fig. 3C), suggesting that the replication of ΔdnaA started from a nearby site. Further, mutations such as in-del or point mutation, plasmid integration and transposition of dnaA gene were not detected in the ΔdnaA clones by high-throughput genomic DNA sequencing. We concluded therefore that dnaA was not essential for the proliferation of Synechococcus sp. PCC 7002 and that the wild-type chromosome replicated in a DnaA-oriC independent manner starting from a specific position.
Our results suggest that loss of DnaA-oriC-dependency independently occurred multiple times during cyanobacterial evolution and raises a possibility that the loss of dnaA or loss of DnaA-oriC dependency correlated with an increase in ploidy level.
Reference
| 1. | Katayama, T., Ozaki, S., Keyamura, K., and Fujimitsu, K. (2010). Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nature Reviews Microbiology 8, 163-170. |
| 2. | Ohbayashi, R., Watanabe, S., Ehira, S., Kanesaki, Y., Chibazakura, T., and Yoshikawa, H. (2016). Diversification of DnaA dependency for DNA replication in cyanobacterial evolution. The ISME journal 10, 1113-1121. |
| 3. | Ohbayashi R, Hirooka S, Onuma R, Kanesaki Y, Hirose Y, Kobayashi Y, Fujiwara T, Furusawa C, and Miyagishima SY. (2020) Evolutionary changes in DnaA-independent chromosomal replication in cyanobacteria. Frontiers in Microbiology. 11:786. |