Latest Research
- 2022.08.01
- Yoshizawa-Sawada Group
Hidden topologies in peptide strands
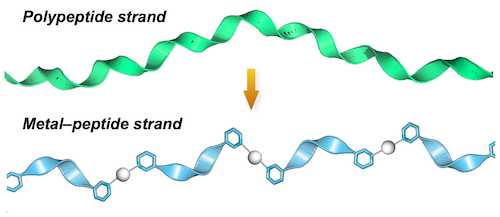
Crochet crafts using flexible string materials are often seen in macroscale, but such a manufacturing method has been undeveloped in microscopic nanoscale. Although phenomena of randomly entangled polymer chains are usually seen, creation of an orderly entangled nanostructure has been difficult. However, in recent years, we have discovered that peptide strands linked by metal ions show a unique property of spontaneously forming a highly entangled nanostructure.[1] We found this phenomenon when trying an idea of extending a short peptide fragment with metal ions (Fig. 1) in our attempts to artificially imitate a protein (polypeptide strands) in nature.
|
|
|
| Fig. 1. Design of a metal-peptide strand. |
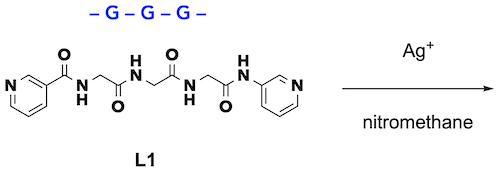
First, we focused on the 3-residue sequence of glycine (G), the simplest amino acid oligomer. 3-Pyridyl groups were introduced at its both ends (ditopic peptide ligand L1), which was complexed with silver ions, a typical linear di-coordinating metal ion, in an organic solvent (Fig. 2).
|
|
|
| Fig. 2. Complexation of tripeptide ligand L1. |
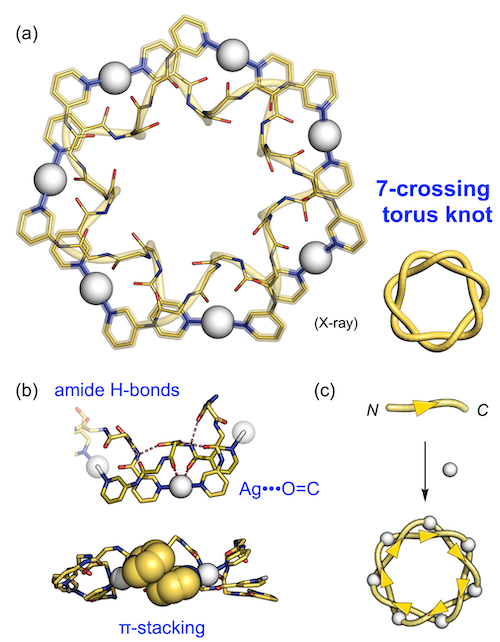
We carried out crystallization from the obtained solution for elucidation of the self-assembled structure. Crystallographic study revealed that the continuous molecular strands of (Ag•L1) formed a unique knotted nanostructure (Fig. 2a).[2] This structure, the double helix closed in a donut shape, has an entangled topology called the "7-crossing torus knot." This was derived from cooperative weak interactions such as amide H-bonds between peptide backbones, π-stacking between pyridine rings, and weak coordination bonds between a silver ion and a peptide carbonyl oxygen (Fig. 2b). Therefore, although ditopic ligand L1 has a directionality from N- to C-termini, all the ligands were perfectly oriented in the knot structure (the perfect head-to-tail manner, Fig. 2c), which indicated remarkable stereoselectivity worked in this self-assembly process.
 |
|
| Fig. 3. | (a) Structure of the 7-crossing torus knot, and (b) various weak interactions and (c) the tacticity found in the knot structure. |
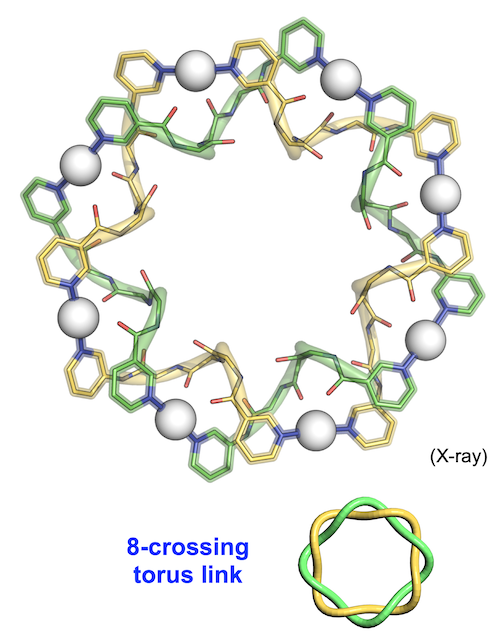
When the solution state was investigated by 1H NMR measurement, another highly symmetrical self-assembled structure was also confirmed. By changing the counter anion of the silver ion used for crystallization, we successfully obtained a single crystal of it and revealed the "8-crossing torus link" structure (Fig. 4). The structural feature that the double helix was closed like a donut was the same as the 7-crossing knot, but it was mathematically classified as not a "knot" (entanglement of one macrocyclic component) but a "link" (that of two or more macrocyclic components) because of the even crossing number. The energy difference between the knot (the Ag7(L1)7 composition) and the link (the Ag8(L1)8 composition) was quite small due to the almost same peptide conformations, which resulted in equilibrium mixture in the solution state.
 |
|
| Fig. 4. Structure of 8-crossing torus link. |
Many attempts have been made to synthesize torus knots/links for many years, but the previous successful examples have been limited to 3- to 6-crossings. Both torus structures in this work became the first synthetic examples of 7- and 8-crossings. Furthermore, since the peptide sequence used here was a simple glycine oligomer, it can be considered that the peptide backbone itself has the latent entangling nature. We consider that the difference from natural polypeptide strands is that reversible coordination bonds are present throughout the molecular strand, allowing them easily to pass through the strand during the self-assembly process.
Next, we tried the peptide sequence of AGG (A: alanine). The 7-crossing knot and 8-crossing link were again obtained from the ligand (L2) by complexation with silver ions. In the crystal structure, it was confirmed that the methyl side chain of A was accumulated in the torus hole (Fig. 5a). Therefore, we considered that the torus structure could be expanded by utilizing the steric hindrance of the side chain in the hole. As expected, 9-crossing torus knot was newly formed from ethyl side chain ligand L3 of the AbuGG (Abu: 2-aminobutyric acid) sequence (Fig. 5b).[3]
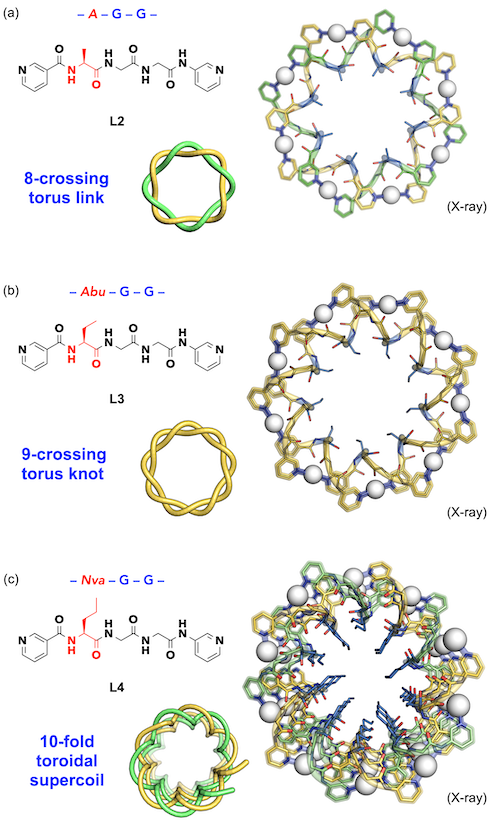 |
|
| Fig. 5. | A series of peptide sequences and crystal structures of advanced torus topologies. (a) An 8-crossing torus link with Me side chains, (b) a 9-crossing torus knot with Et side chains, (c) a 10-fold toroidal supercoil with n-Pr side chains. |
Furthermore, when n-propyl ligand L4 of the NvaGG (Nva: norvaline) sequence was examined, it was suggested that a 10-crossing torus link was formed in solution state. However, the crystal structure was an unexpected nanostructure: a toroidal supercoil structure (infinite polymeric structure) emerged (Fig. 5c). This is a supercoiled structure in which 10 units of double helical (Ag•L4) strands formed one pitch of larger helix, which could be generated by the ring opening of the 10-crossing torus link and connected infinitely. It can be expressed as a "supramolecular ring-opening polymerization" triggered by steric strain.
In summary, we successfully revealed that metal-peptide strands yielded highly entangled nanostructures. In nature, DNA circular plasmids and supercoils are well known as highly entangled nanostructures. When DNA forms entangled nanostructure, topoisomerase, an enzyme that works for temporary cutting and recombining of the strand, enables strands threading for an ideal entangled nanostructure. In this metal-peptide strand system, the reversible coordination bond plays such an important role, and thus efficiently construct entangled nanostructures. Not only the simple glycine-based peptide sequences introduced here, but proline-rich sequences can give another type of highly entangled nanostructure of even larger crossing numbers (called as polyhedral links).[4,5] Elucidation of structural dynamics of such entangled nanostructures and utilization of nanospace having a topological chirality will be reported in the future.
| [1] | Bull. Chem. Soc. Jpn. 2021, 94, 2342. (https://doi.org/10.1246/bcsj.20210218) |
| [2] | Chem 2020, 6, 294. (https://doi.org/10.1016/j.chempr.2019.12.009) |
| [3] | J. Am. Chem. Soc. 2021, 143, 16734. (https://doi.org/10.1021/jacs.1c08094) |
| [4] | Nat. Commun. 2019, 10, 921. (https://www.nature.com/articles/s41467-019-08879-7) |
| [5] | Nat. Commun. 2019, 10, 5687. (https://www.nature.com/articles/s41467-019-13594-4) |