Latest Research
- 2022.10.01
- Tanaka-Yoshida Group
Biosynthesis and regulation of a novel sulfated polysaccharide, synecan, in the unicellular cyanobacterium Synechocystis sp. PCC 6803.
【Backgrounds】
Cyanobacteria (blue-green algae), the oxygenic photoautotrophic bacteria, are important organisms that supply oxygen and organic compounds to the environment. In these days, they have been attracting attention as host organisms that utilize their photosynthetic capabilities to produce substances with minimal environmental impact. In nature, cyanobacteria inhabit a wide range of environments, such as lake, stream, hot spring, ocean, and land, and form a variety of biofilms like microbial mat, colony, and bloom (water bloom). One of the major components of cyanobacterial biofilm is the extracellular polysaccharides (EPS) they produce. However, the biosynthesis and regulation mechanisms of cyanobacterial EPSs and their physiological functions remain unrevealed. So, we have been working to figure it out1.
Cyanobacteria make a wide variety of EPSs, of which sulfated polysaccharides are characteristic.2。Sulfated polysaccharides are polysaccharides modified with sulfate groups, and many of them are known to have the water retention property and viscosity. Naturally, they exist as glycosaminoglycans in animals (e.g., chondroitin sulfate), cell wall polysaccharides in eukaryotic algae (e.g., carrageenan), and EPSs in cyanobacteria. Interestingly, almost no bacteria other than cyanobacteria are known to produce sulfated polysaccharides. On the other hand, sulfated polysaccharides of various compositions are present in cyanobacteria and are thought to be involved in the formation of biofilm such as colonies and in physiological functions. In addition, sulfated polysaccharides derived from cyanobacteria, such as spirulan from Spirulina (Arthrospira platensis) and sacran from Suizenji-nori, are being studied as potential materials for pharmaceuticals, cosmetics, and fibers3. Thus, sulfated polysaccharides of cyanobacteria are important EPS in terms of both basic and applied research. However, their biosynthesis and regulation mechanisms had not been elucidated.
In this study, we discovered a novel cyanobacterial sulfated polysaccharide, synechan, and identified a comprehensive set of genes involved in its biosynthesis and regulation4. Since detailed data are provided in the paper, only excerpts are presented in the text.
【Results】
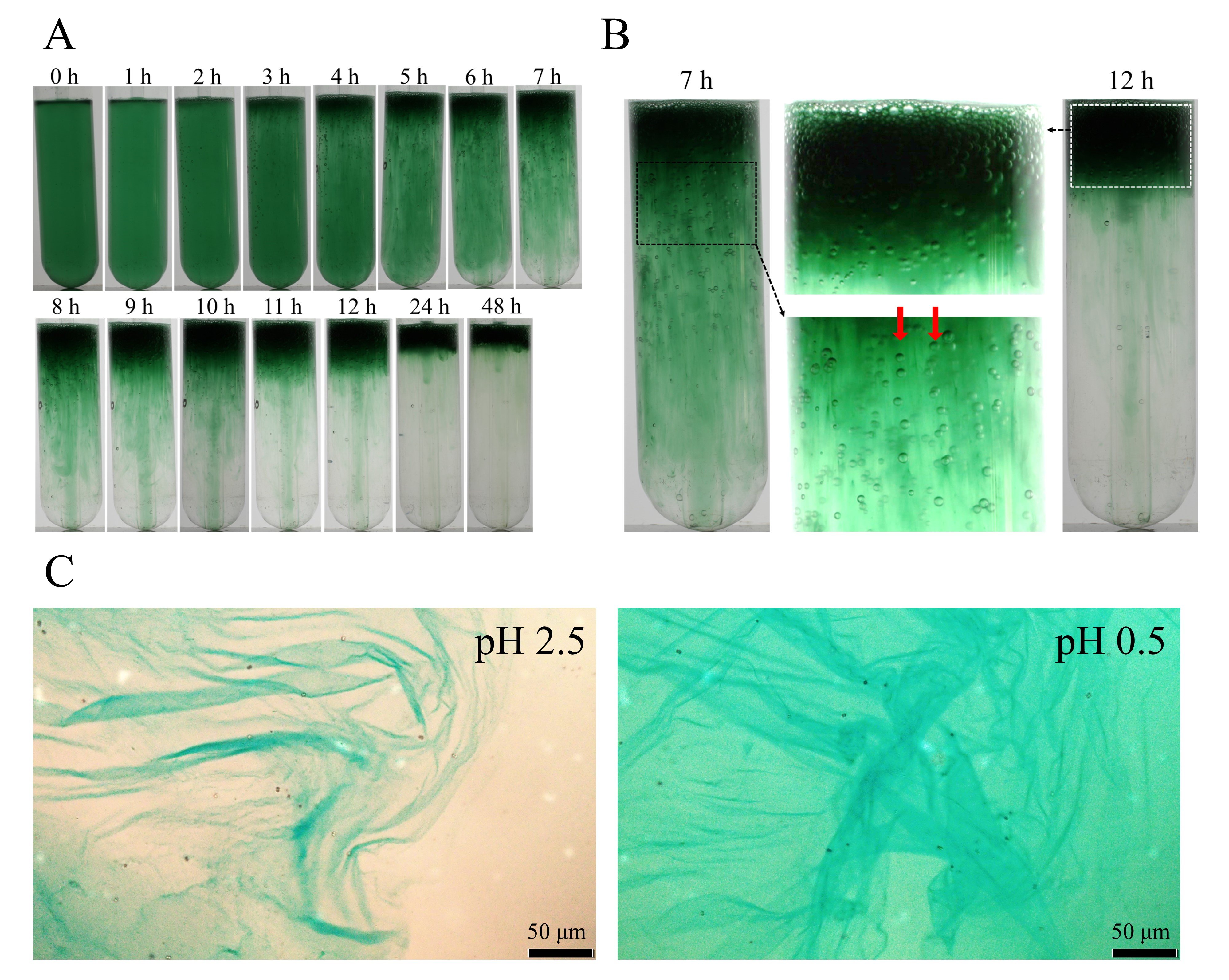
To identify biosynthesis genes of cyanobacterial sulfated polysaccharide, a species that can synthesize a sulfated polysaccharide and can be transformed was needed. First, we found that one of the substrains of Synechocystis sp. PCC 6803 (hereafter S.6803), a unicellular cyanobacterium that has long been used as a model organism for research, formed a viscous cell mass at the liquid surface when the culture medium was standing (Fig. 1A, B, Movie 1). We also found that this viscous material was EPS and contained slfated polysaccharides by Alcian blue staining and chemical composition analysis (Fig. 1C), and we designated the polysaccharide as synecan.。
|
Movie 1. The viscous cell aggregate of S.6803.
|
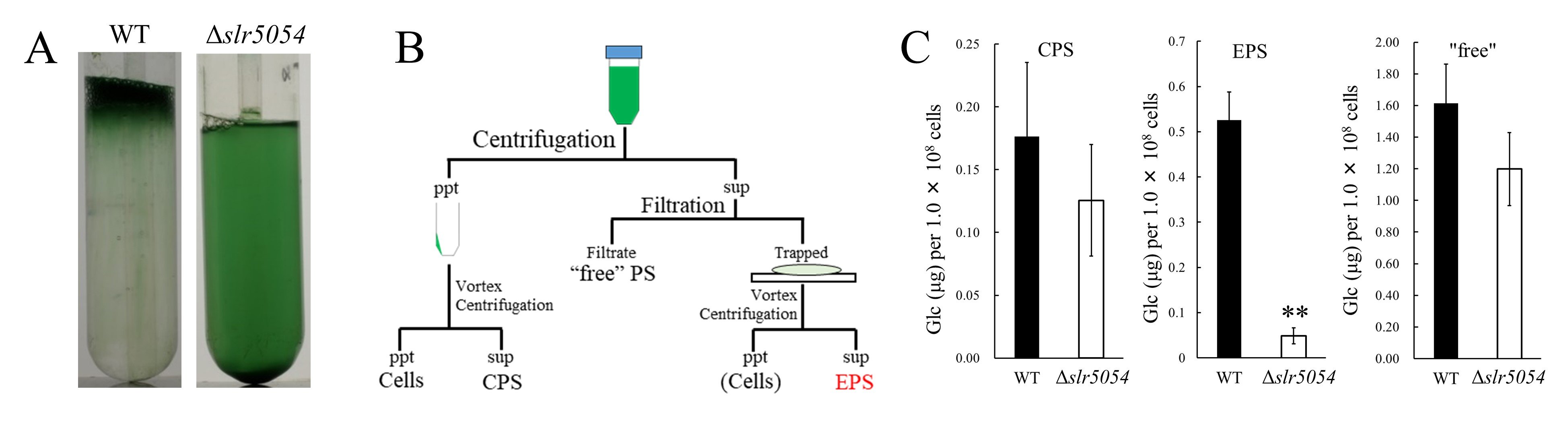
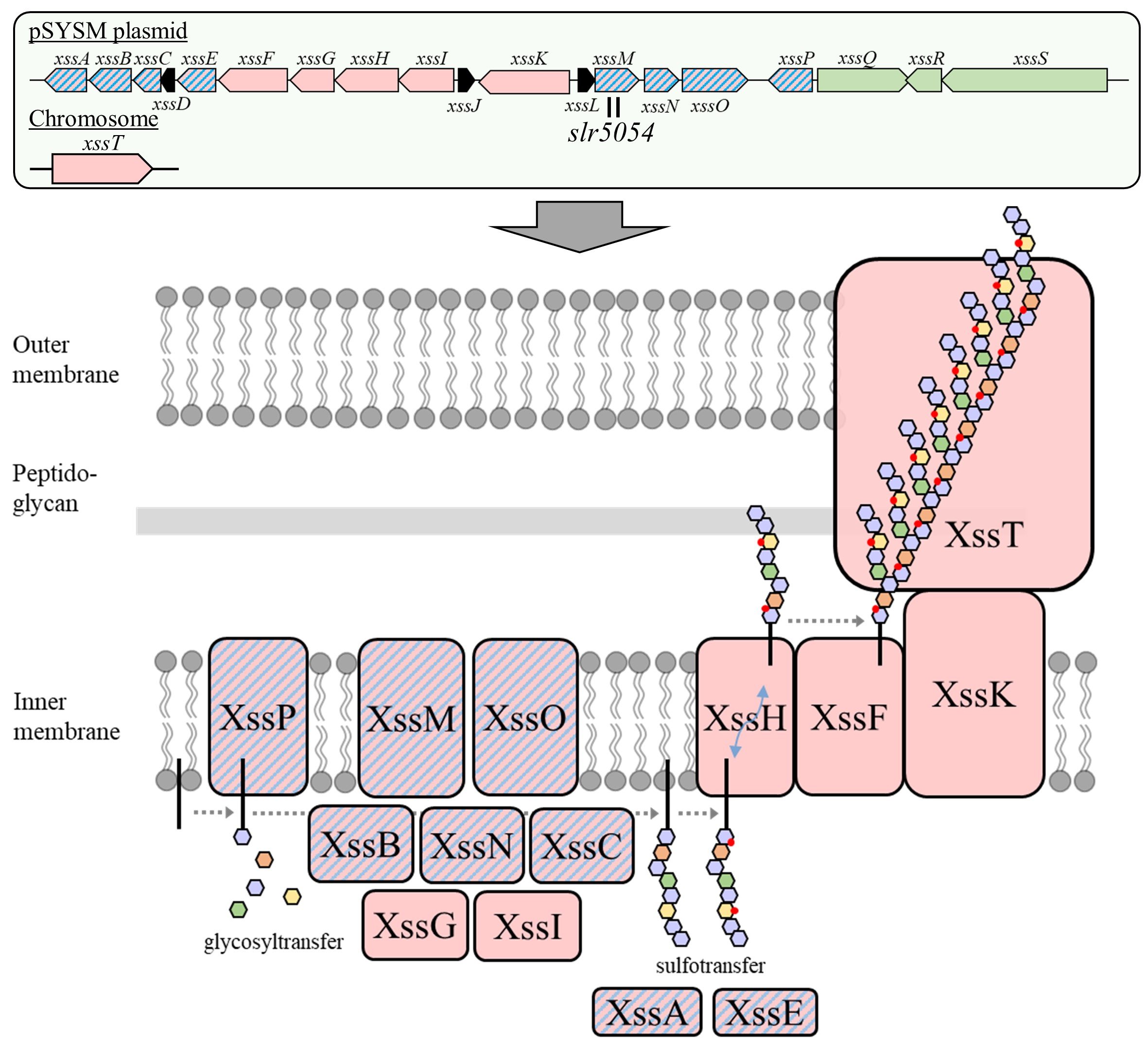
Next, we identified the synecan biosynthesis genes. Bacterial EPS biosynthesis systems are divided into three major types based on their biosynthesis mechanisms and proteins, all of which require transmembrane glycosyltransferases5. Therefore, we generated several disruption mutants of the "transmembrane glycosyltransferase" in S. 6803 and analyzed their phenotypes (Fig. 2A). As a result, only the disruption mutant of slr5054 (Δslr5054) did not form the viscous aggregate. In addition, when viscous EPS, cell surface polysaccharides (CPS), and low molecular weight EPS were fractionated and quantified from wild type (WT) and slr5054 disruptant (Δslr5054) mutant respectively, only the amount of viscous EPS fraction was significantly reduced in Δslr5054 (Fig. 2B and C). These results indicated that slr5054 was one of the genes required for synecan biosynthesis. slr5054 was located within a large gene cluster on the mega plasmid pSYSM of S.6803 (Fig. 3). The cluster consisted of a total of 19 genes, including the Wzx/Wzy-type EPS biosynthesis genes, sulfotransferase genes, and regulatory genes to be described later. Therefore, we constructed disruption mutants of individual genes and analyzed their phenotypes and found that most of them were required for synechan biosynthesis. We designated these synechan biosynthesis genes as xss (extracellular sulfated polysaccharide biosynthesis).
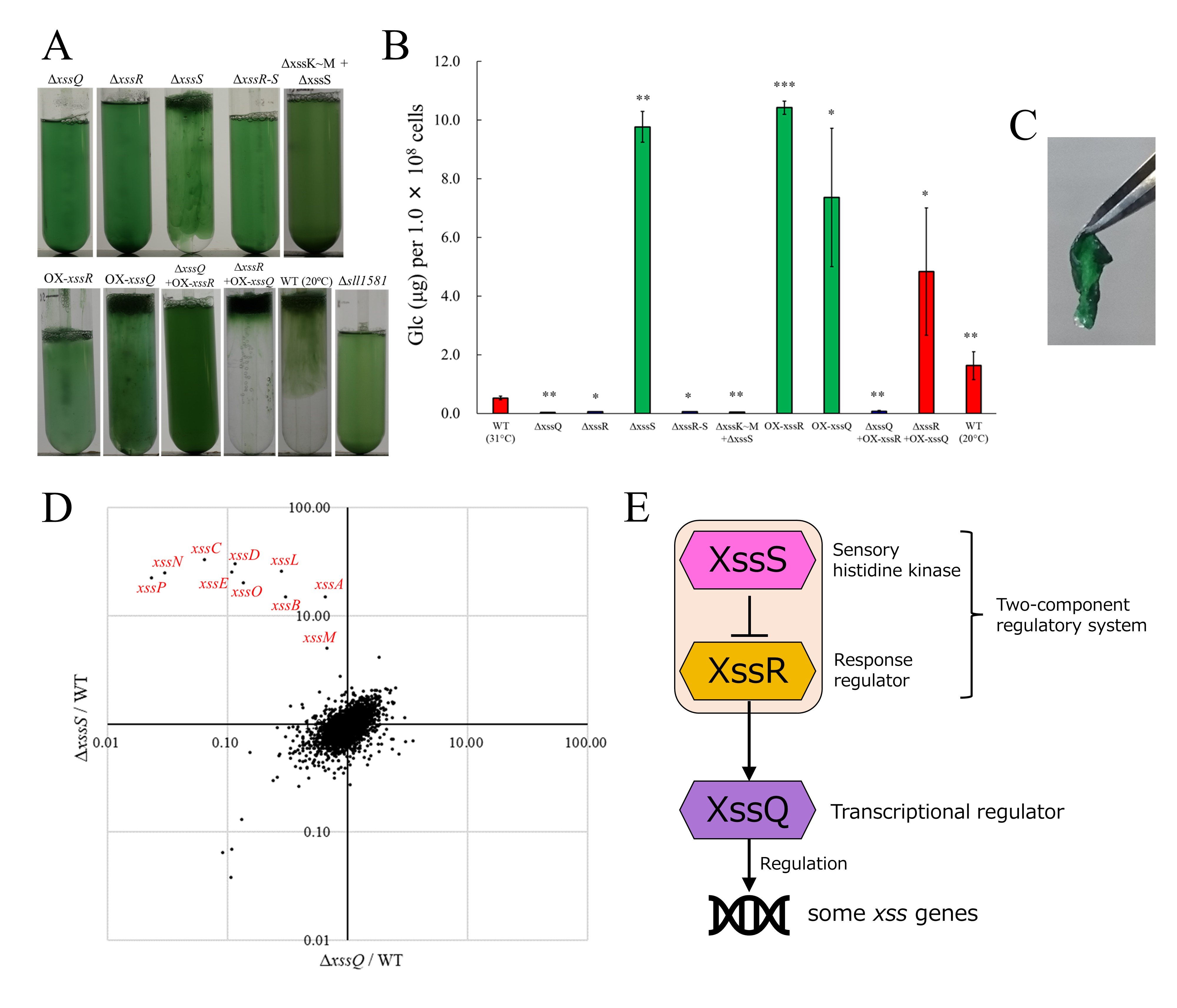
There also existed within the xss gene cluster on pSYSM a transcriptional regulator-like gene, xssQ, a sensor histidine kinase gene, xssS and a response regulator gene, xssR. The disruption strains of xssQ and xssR (ΔxssQ and ΔxssR) did not form viscous cell aggregates and accumulated little EPS (Fig. 4A, B). On the other hand, in the xssS disruptant mutant (ΔxssS), the entire culture medium resembled a viscous cell aggregate and EPS accumulation increased to about 19 times that of WT. Overexpressing strains of xssQ and xssR (OX-xssQ and OX-xssR) also showed the same phenotype as ΔxssS and formed sheet-like biofilm on agar plate (Fig. 4C). Comparing the transcript levels of genes based on RNA-seq analysis between WT, ΔxssQ, and ΔxssS, we found that genes with decreased transcription in ΔxssQ and increased in ΔxssS compared to WT, in other words, genes that were positively correlated with synechan biosynthesis ability, were only a part of the xss genes (Fig. 4D). These results indicate that XssSRQ regulates synechan biosynthesis via transcriptional control of some xss genes and that XssS acts in a repressive manner on the biosynthesis.
【Discussion】
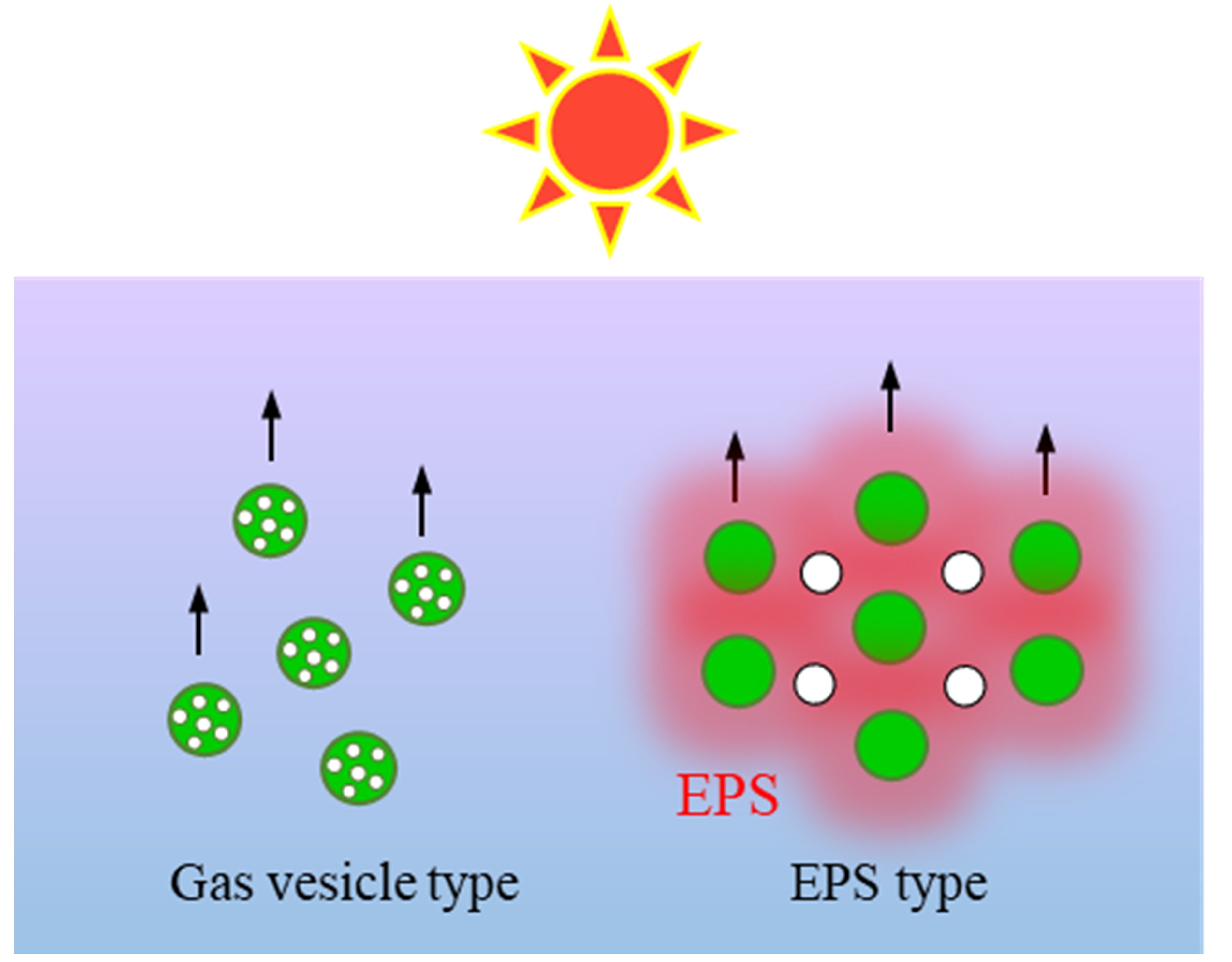
What does S.6803 make synechan for? We consider bloom formation as one possibility. Cyanobacteria are known to float to the surface of the water, a light-rich environment, and form blooms, and their buoyancy is mainly provided by intracellular gas vesicles. On the other hand, S.6803 formed bloom-like viscous cell aggregates by buoyancy of bubbles trapped in extracellular synechan, despite lack of gas vesicles. Based on this, we propose that a new mechanism for cyanobacterial bloom formation may be due to extracellular EPS and gas bubbles (Fig. 5). However, this is still hypothetical. In addition, the details of the transcriptional regulation mechanism by XssSRQ and the environmental stimuli that XssSRQ senses are still unclear. We would like to further our understanding of the function of synechan and the ecology of S.6803 by studying these in the future. This study is the first comprehensive elucidation of the genes involved in the biosynthesis and regulation of sulfated polysaccharides in cyanobacteria, and the results are expected to contribute to the study of sulfated ones in other cyanobacteria. For example, we could estimate the genes related to sulfated polysaccharide biosynthesis by searching the genomic information of other cyanobacteria based on the information of the xss genes. In addition, xssSRQ and especially xssQ are present in the genome of certain number of cyanobacteria and are assumed to serve as a regulatory system for gene expression in those species as well.
The outcome of this research is also expected to be utilized for applied purpose. As mentioned in the background, cyanobacteria can fix CO2 through photosynthesis to produce a variety of substances, including EPS. And cyanobacterial sulfated polysaccharides are expected to be industrially useful substances and are being studied. Therefore, as we were able to improve synechan productivity by disrupting xssS, we would like to promote this study to research on modifying the productivity and composition of sulfated polysaccharides by genetic modification. Moreover, whether synechan is an industrially useful polysaccharide like other cyanobacterial sulfated polysaccharides is largely unexplored at this time and will be clarified in the future.
| 1. | Maeda K, Tamura J, Okuda Y, Narikawa R, Midorikawa T, Ikeuchi M. 2018. Genetic identification of factors for extracellular cellulose accumulation in the thermophilic cyanobacterium Thermosynechococcus vulcanus: proposal of a novel tripartite secretion system. Molecular Microbiology 109:121-134 |
| 2. | Pereira S, Zille A, Micheletti E, Moradas-Ferreira P, De Philippis R, Tamagnini P. 2009. Complexity of cyanobacterial exopolysaccharides: composition, structures, inducing factors and putative genes involved in their biosynthesis and assembly. FEMS Microbiology Reviews 33:917-941 |
| 3. | Okajima MK, Bamba T, Kaneso Y, Hirata K, Fukusaki E, Kajiyama Si, Kaneko T. 2008. Supergiant ampholytic sugar chains with imbalanced charge ratio form saline ultra-absorbent hydrogels. Macromolecules 41:4061-4064 |
| 4. | Kaisei Maeda, Yukiko Okuda, Gen Enomoto, Satoru Watanabe, Masahiko Ikeuchi. Biosynthesis of a sulfated exopolysaccharide, synechan, and bloom formation in the model cyanobacterium Synechocystis sp. strain PCC 6803. eLifei, 2021, 10:e66538. |
| 5. | Schmid J, Sieber V, Rehm B. 2015. Bacterial exopolysaccharides: biosynthesis pathways and engineering strategies. Frontiers in microbiology 6:496. DOI: 10.3389/fmicb.2015.00496, PMID: PMC4443731 |