Latest Research
- 2023.08.01
- Kitaguchi Group
Patrol Yeast: A new cell-based biosensor system detecting toxic substances associated with food poisoning
Toxins, viruses, and bacteria in food present a considerable threat to human well-being. Aflatoxins, which are generated by fungi in deteriorated food, are extremely carcinogenic and can result in liver cancer. Raw foods can be contaminated with enterohemorrhagic bacteria like E. coli O111 and O157, causing severe symptoms in individuals. Existing approaches to analyzing these contaminants involve costly equipment and skilled personnel, making them inaccessible in numerous contexts. Therefore, there is a pressing requirement for a quick, convenient, and cost-effective analytical method to mitigate the risk of foodborne illnesses, particularly in developing nations.
Previously, mammalian cell-based biosensors using antibody-based receptors have been developed by other groups [1] and showed promise in the analysis field. However, there is a lack of such biosensors using baker's yeast cells, which can be propagated easily in low-cost and well-suited for withstanding harsh environments, while safe for human consumption. The utilization of yeast cells poses a challenge due to the hindrance caused by their cell wall, which obstructs the access of large analytes to the detection unit on the yeast membrane. In this study, through partially enzymatic digestion of the cell wall, we developed a baker's yeast-based biosensor system called "Patrol Yeast" (Fig. 1) that combines antibody receptors with a membrane-based yeast two-hybrid system (MbY2H) [2]. This system allows for the detection of various substances, from small molecules to microorganisms, and has the potential to identify and monitor food contaminants effectively.
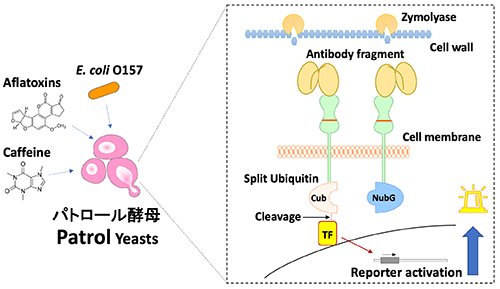 |
|
| Fig. 1. | Diagram of the detection system by Patrol Yeast [2]. Analytes not passing through the cell walls of yeast become accessible to recognition units (antibody fragments) at optimal concentrations of Zymolyase. Cleaved transcription factor (TF) by split ubiquitin complementation activates the reporter enzyme gene, which is brought about by analyte recognition. NubG, N-terminal of ubiquitin with I13G mutation; Cub, C-terminal region of ubiquitin. |
We first developed Patrol Yeast biosensors to detect specific small molecule targets. We successfully detected a model antigen called Bone Gla Protein (BGP) and demonstrated the feasibility of detecting small molecules such as caffeine (food additive) and aflatoxins (toxic metabolite from fungi contaminations) (Fig. 2). By utilizing antibody fragments as affinity agents, including the open-sandwich antibody fragments pairs (Fig. 2A) or antigen-triggered VHH dimerization, the Patrol Yeast biosensors were able to detect these targets through the activation of reporter genes, β-galactosidase (β-Gal), which provided a measurable signal after a simple overnight co-incubation with the samples to be analyzed.
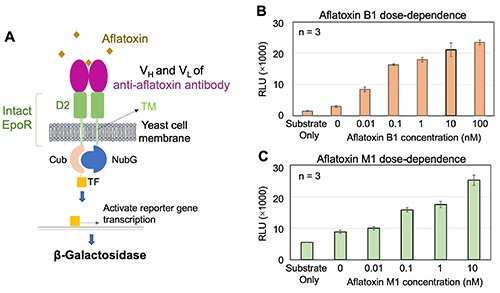 |
|
| Fig. 2. | Aflatoxins were detected by Patrol Yeast [2]. A, Mechanism by which Patrol Yeast detects aflatoxin. B, C, Aflatoxin B1 and Aflatoxin M1 were detected in a dose-depend manner. Data are shown as means ± standard deviation. D2, D2 domain of erythropoietin receptor (EpoR); VH, variable region of heavy chain; VL, variable region of light chain; TM, transmembrane domain. |
The biosensors showed dose-dependent responses to aflatoxins, with low limits of detection (Fig. 2B-C). We also extended the Patrol Yeast's ability to detect enterohemorrhagic E. coli O157, a larger microorganism target. By optimizing the partial digestion of yeast cell walls using an enzyme mix called Zymolyase, the biosensors were able to detect E. coli O157 in a dose-dependent manner (Fig. 3).
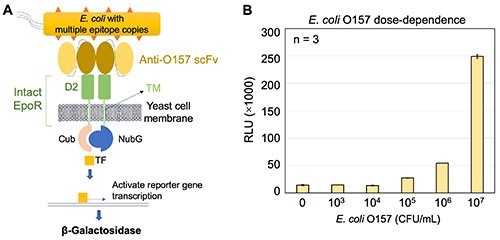 |
|
| Fig. 3. |
E. coli O157 was detected by Patrol Yeast [2]. A, Mechanism by which Patrol Yeast detects E. coli O157. B, E. coli O157 was detected in a dose-dependent manner. Data are shown as means ± standard deviation. scFv, single-chain variable fragment of an antibody.
|
To further expand the capabilities of the Patrol Yeast biosensors, we integrated a secreted luciferase reporter system. This system allowed for the detection using a simple luminescence assay, enhancing both the convenience and sensitivity of the Patrol Yeast (Fig. 4).
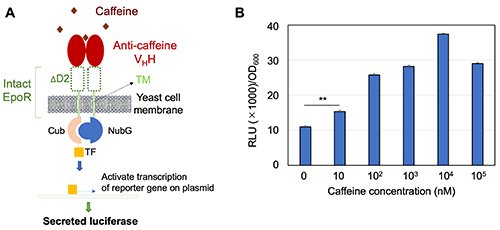 |
|
| Fig. 4. |
Caffeine was detected by secreted CLuc from Patrol Yeast [2]. A, Mechanism by which Patrol Yeast with secreted CLuc reporter detects caffeine. B, Caffeine activated the secretion of CLuc luciferase from Patrol Yeast in a dose-dependent manner. *Two-tailed Welch t-tests, p < 0.005. Data are shown as means ± standard deviation. ΔD2, removed EpoR D2 domain.
|
The Patrol Yeast biosensor system presents an innovative and practical solution for identifying toxins and microorganisms in food and beverages. In comparison to alternative analytical methods, this system offers affordability, convenience, and ease of use, thanks to the well-developed technology for large-scale yeast production. This differentiates it from mammalian cell-based sensors that necessitate expensive media and meticulous culture in a humid environment. The biosensors demonstrated good sensitivity and met food contamination regulations for caffeine and aflatoxins detection. The β-Gal reporter from the original MbY2H system requires a cell disruption process before the signal detection. The inclusion of a secreted luciferase reporter (CLuc) alongside the established β-Gal reporter further enhanced the feasibility of the Patrol Yeast system by minimizing the risk of aerosol generation and reducing equipment requirements during practical operations. In summary, we firmly believe that the Patrol Yeast system will make a valuable contribution to the field of food safety, serving as a sensitive, cost-effective, and user-friendly tool for detecting toxins and monitoring a broad range of targets.
| [1] | Manhas, J., Edelstein, H.I., Leonard, J.N., Morsut, L., 2022. Nat. Chem. Biol. 18, 244-255. DOI: 10.1038/s41589-021-00926-z |
| [2] | Su, J.#, Zhu, B.#, Inoue, A., Oyama, H., Morita, I., Dong, J., Yasuda, T., Sugita-Konishi, Y., Kitaguchi, T., Kobayashi, N., Miyake, S., Ueda H., 2023. Biosens. Bioelectron., 219, 114793. #Equal contributions. DOI: 10.1016/j.bios.2022.114793 |



