Latest Research
- 2024.10.01
- Tanaka-Yoshida Group
Novel regulatory mechanism of GTP synthesis involved in the adaptation for amino acid starvation in bacteria
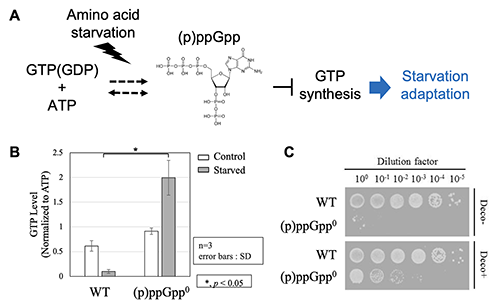
Cells have a variety of energy molecules that support biological activities. Among them, guanosine triphosphate (GTP) is an energy molecule used as energy currency to drive "dynamic" cellular biological processes such as protein synthesis and cell division and plays a role in determining the level of cell activity. The amount of intracellular GTP in bacteria varies greatly with growth phase, and its amount correlates with doubling time of the cell. One cellular response that is particularly well characterized by GTP is the nutrient starvation response. When bacteria are exposed to an environment depleted of nutrient sources, they arrest proliferation and synthesize the nutrient sources essential for survival. It is known that a decline in GTP synthesis is important for this metabolic switch1. The mechanism responsible for regulating GTP synthesis is known as the "stringent response". In the stringent response, small nucleotide molecules (p)ppGpp are synthesized from GTP (GDP) in response to stresses such as amino acid starvation. (p)ppGpp directly inhibits GTP biosynthesis resulting in decreasing GTP levels. As a result, growth is repressed while expression of genes involved in amino acid biosynthesis is promoted, allowing adaptation to amino acid starvation2,3 (Figure 1A). Although it has been shown that (p)ppGpp-mediated regulation of GTP synthesis is essential for adaptation to amino acid starvation conditions, what factors besides (p)ppGpp are involved in the regulation of GTP synthesis is not well understood.
In Bacillus subtilis, a strain lacking all genes encoding the enzyme responsible for (p)ppGpp synthesis [(p)ppGpp0 strain] shows growth inhibition under the amino acid starvation conditions. In the wild-type strain, GTP levels decrease when amino acid starvation is induced, whereas GTP levels increase in the (p)ppGpp0 strain (Figure 1B). The growth inhibition of the (p)ppGpp0 strain under amino acid starvation condition is suppressed by inhibitors or gene mutations of enzymes in the GTP synthesis pathway (Figure 1C). These findings suggest that the inability to inhibit GTP synthesis is a factor in the growth inhibition of the (p)ppGpp0 strain under amino acid starvation conditions. We hypothesized that genetic analysis of this (p)ppGpp0 strain would reveal a new factor involved in the regulation of GTP synthesis, which is essential for amino acid starvation condition. In this study, we found that methionine metabolism is involved in the regulation of GTP synthesis, which is essential for adaptation to amino acid starvation, by using this (p)ppGpp0 strain4.
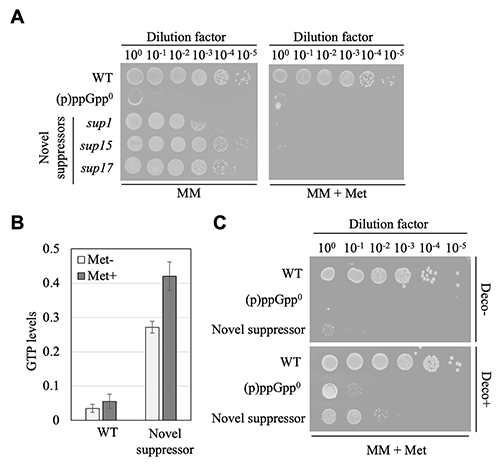
We first performed genetic screening of suppressor mutants that allowed (p)ppGpp0 strain grow under amino acid starvation medium conditions. Previous studies performed screened similar genetic screening and identified mutations in genes encoding enzyme responsible for GTP synthesis pathway as the causative mutation that allowed suppressor mutants to recover growth5. Through our screening, we identified suppressor mutants without mutations in the genes encoding enzyme responsible for GTP synthesis pathway in addition to the previously screened suppressor mutants with mutations in the genes encoding enzyme responsible for GTP synthesis pathway. Further analysis of the "novel suppressor mutants" without mutation in the genes encoding enzyme responsible for GTP synthesis pathway revealed that growth recovery of novel suppressor mutants reverted when methionine was added to the amino acid starved conditions (Figure 2A). On the other hand, the suppressor mutants, which has a mutation in the genes encoding enzyme responsible for GTP synthesis pathway, showed no growth inhibition effect upon addition of methionine, suggesting that methionine may somehow promote GTP synthesis and cause growth inhibition in the novel suppressor mutants. As expected, the intracellular GTP levels were markedly increased by methionine addition in the novel suppressor mutant (Figure 2B). The growth inhibition by methionine addition was complemented by the addition of inhibitors of enzymes in the GTP synthesis pathway (Figure 2C). These results suggest that growth recovery of novel suppressor mutants reverted as a result of enhanced GTP synthesis upon methionine addition.
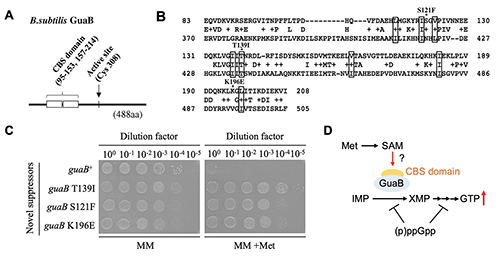
We then explore the mechanism by which methionine promotes GTP synthesis. We analyzed genetically which step of the GTP synthesis pathway methionine promotes and found that the effect of methionine may originate from the enzyme IMP dehydrogenase (GuaB), which is responsible for the initial reaction in the IMP to GTP biosynthesis pathway. GuaB has a conserved subdomain called the CBS domain, which is known to bind guanine and adenine nucleotides as a ligand (Figure 3A). The CBS domain is also present in enzymes other than GuaB, mainly as a domain to which compounds with an adenine backbone bind. S-adenosylmethionine (SAM) is synthesized from methionine by addition of the adenine backbone of ATP. There are several enzymes that bind this SAM as a ligand for the CBS domain. The crystal structure of SAM binding to the CBS domain of an Archaeal enzyme has been solved, and we noted that the amino acid residues involved in SAM binding identified there are conserved in the CBS domain of B. subtilis GuaB in almost the same form (Figure 3B). When mutations of amino acid residues located near these residues were introduced into a novel suppressor mutant, growth inhibition by methionine was suppressed (Figure 3C). These results suggest that SAM synthesized from methionine may activate GuaB through the CBS domain and promote GTP synthesis (Figure 3D).
Methionine is known to be a particularly important amino acid for protein synthesis. Its metabolite, SAM, is also an important metabolite which plays a role as a major donor of methyl groups in methylation modifications of various biomolecules. Our study has revealed for the first time that methionine metabolism, which supports these vital biological reactions, directly regulates GTP biosynthesis.
It has been revealed that the regulation of GTP synthesis is important for regulating cell growth not only in bacteria but also in mammalian cancer cells6. In other words, the regulation of GTP synthesis is thought to play an important role in the control of growth in all organisms. While the regulation of GTP synthesis by (p)ppGpp and the feedback regulation of GTP to the rate-limiting enzyme have been clarified in bacteria, other factors involved in GTP synthesis are not well understood in all organisms. The challenge for the future, from bacteria to humans, will be to clarify how GTP synthesis is regulated by factors depending on the state of cell proliferation. We believe that this will lead to an understanding of the common rules of the system by which cells regulate proliferation and the diversification process during the process of biological evolution.
<References>
| [1] | Lopez., Arch Microbiol., 131(3):247-51 (1982) |
| [2] | Krásný & Gourse, EMBO J., 23(22):4473-83 (2004) |
| [3] | Krásný et al., Mol. Microbiol., 69(1):42-54 (2008) |
| [4] | Osaka et al., Mol. Microbiol., 113(6):1155-1169 (2020) |
| [5] | Kriel et al., Mol. Cell, 48, 231-241 (2012) |
| [6] | Kofuji et al., Nat. Cell Biol., (8):1003-1014 (2019) |