Latest Research
- 2015.04.30
Understanding of The Genome-wide Transcriptional Hierarchy of Bacteria
by Genomic SELEX
1.Introduction
Organisms monitor extracellular environmental conditions, and can respond by selecting their genome expression profiles. Regulation of gene expression is in general mainly determined by the transcription step. The gene selectivity of RNA polymerase is modulated after interaction with two groups of regulatory proteins, sigma factors and transcription factors (ref. 1). Recently, the whole genome sequence of various organisms has been determined by advanced sequencing technology, and thus far massive genes have been annotated. However, it is difficult to identify a regulation of gene sets by available genome information, and therefore, to elucidate the regulation system of genome transcription is one of the research frontiers.
2.The necessity of identification of direct targets of transcriptional regulators
As hitherto methods for analysis of transcription regulation, it is useful to compare the gene expression patterns between the parental wild-type strain and the defective mutant of the target regulator. Recently, based on the revealed genome information, comprehensive analysis methods such as transcriptome analysis and proteome analysis has been established. While these comprehensive technologies could identify any effects by the defect of the target transcription regulator, it is difficult to distinguish direct from indirect regulations. Furthermore, it has been proposed that transcription regulators are composing regulatory network, which is likely constituting a transcriptional hierarchy (ref. 2). Therefore, in order to understand the molecular mechanism of genome transcriptional regulation system, it was necessary to develop a technology to identify the direct target genes of transcription regulators.
3.Genomic SELEX analysis
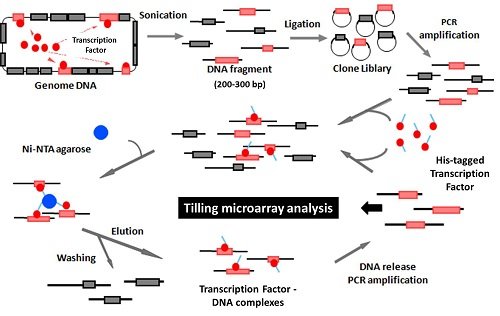
To identify the direct target genes of transcription regulators, we developed a new method called Genomic SELEX analysis (ref. 3). In this method, DNA-protein complexes were isolated from the mixture of any DNA-binding protein and the fragmented genome DNA (Fig. 1).
Figure 1. Genomic SELEX system
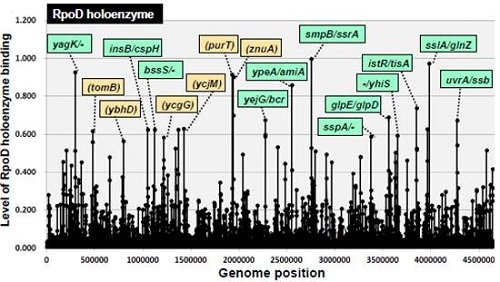
For determination of the sequences of protein-bound SELEX DNA fragments, we use cloning and sequencing procedures at first, and then use tilling array procedures for comprehensive analysis at the genome level (ref. 4). Using this improved method, we succeeded to identify the binding sites of RNA polymerase on the E. coli genome (Fig. 2,ref. 5, 6).
Figure 2. Distribution of RNA polymerase holoenzyme (Sigma70) on E.coli genome
This experiment was done with the RNA polymerase core enzyme containing the principal sigma factor sigma-70 and the E. coli genome DNA. Thus, it was expected that identified DNA binding sites included promoters are recognized by sigma-70 holoenzyme alone. Thus far reported promoter sequences determined based on in vivo analyses likely include promoters that require some additional regulatory factors for activation. Here, we proposed promoters selected by Genomic SELEX as 'Constitutive promoter', and the latter promoter as 'Inducible promoter'. In this way, the Genomic SELEX method can provide a tool for the identification and mapping of direct targets on the genome, and it should be a powerful method to elucidate the transcription hierarchy.
4.Future works
In addition to previous comprehensive in vivo analyses involved with any factors, it is critical to make clear the network of genome transcription regulation by the identification of direct targets of each transcription regulator. Based on such knowledge, it is possible to understand the effect along with the time path, because intracellular genome transcription regulation shows dynamic variations. We would like to reveal the intracellular dynamics of transcriptional regulation and the hierarchy structure for the genome-wide transcriptional regulation.
References
1.Ishihama, A. Functional modulation of Escherichia coli RNA polymerase. Annu. Rev. Microbiol. 54, 499-518. (2000)
2.Martinez-Antonio, A. and Collado-Vides, J. Identifying global regulators in transcriptional regulatory networks in bacteria. Curr. Opin. Microbiol.6, 482-9. (2003)
3.Shimada, T., Fujita, N., Maeda, M. and Ishihama, A. Systematic search for the Cra-binding promoters using genomic SELEX system. Genes Cells10, 907-18.
4.Shimada, T., Yamamoto, K., Fujita, N. and Ishihama, A. Novel members of the cAMP receptor protein (CRP) regulon for transport and metabolism of carbon sources. PLoS ONE6, e20081. (2011)
5.Shimada, T., Yamazaki, Y., Tanaka, K. and Ishihama, A. The whole set of constitutive promoters recognized by RNA polymerase RpoD holoenzyme of Escherichiacoli. PLoS ONE9, e90447. (2014)
6.Conway, T., Creecy, J. P., Maddox, S. M., Grissom, J. E., Conkle, T. L., Shadid, T. M., Teramoto, J., San, Miguel, P., Shimada, T., Ishihama, A., Mori, H. and Wanner, B. L. Unprecedented high-resolution view of bacterial operon architecture revealed by RNA sequencing. MBio. 5, e01442-14. (2014)